Fig. Nuclei are the toughest intracellular organelles. When aligned, they act like structural pillars supporting the structure of the gut and influencing asymmetrical changes in shape during its development.
From symmetry to asymmetry: the two sides of life
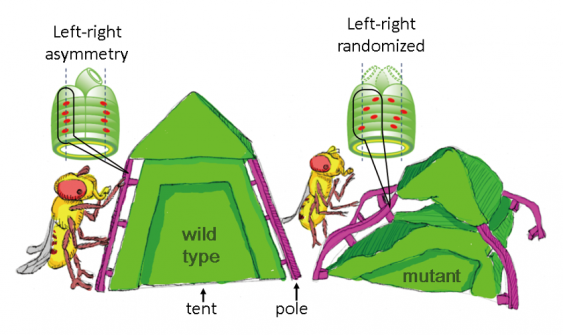
On the outside, animals often appear bilaterally symmetrical with mirror-image left and right features. However, this balance is not always reflected internally, as several organs such as the lungs and intestines are left-right (LR) asymmetrical. Researchers at Osaka University, using an innovative technique for imaging movement of cell nuclei in living tissue, have determined the patterns of nuclear alignment responsible for LR-asymmetrical shaping of internal organs in the developing embryo.
Embryogenesis involves complex genetic and molecular processes that transform a single-celled zygote into a complete, living individual with multiple functional axes, including the LR axis. A long-standing conundrum of Developmental Biology is the breaking of LR symmetry in the developing embryo to initiate lateralization (dominance of one side over the other) of organs and other structures of the body along the LR axis. Mechanisms that induce LR asymmetry are well known in vertebrates; however, the biomechanics in invertebrates remain uncertain.
The research team focused on the development of the intestine, in particular the anterior midgut (AMG) of the Drosophila (fruit fly) as an appropriate model to study LR-asymmetric development in invertebrates. “Proper organ development often requires nuclei to move to a specific position within the cell,” first author Dongsun Shin explains. “Our previous studies had revealed the molecular signaling pathways as well as the proteins responsible for the biomechanical control of asymmetric development, but the dynamics had not yet been determined.”
To meet the challenge of dynamically tracking nuclei in living tissue, the researchers innovated a new method to investigate nuclear migration. Using a confocal laser scanning microscope, they obtained three-dimensional (3D) time lapse videos of immunostained Drosophila midgut at the appropriate embryonal stage. Then, applying mathematical modeling and computational imaging techniques they generated 3D-surface animated models of nuclear migration in the visceral muscles.
Through 3-D time-lapse movies, the researchers identified the initial LR-symmetrical distribution of AMG muscle nuclei along the antero-posterior (front-to-back) axis – termed ‘proper nuclear positioning’. Further, they vividly imaged ‘collective nuclear behavior’ in which crowded nuclei actively rearranged their relative positions with LR symmetry. This symmetric initial ‘proper nuclear positioning’ and ‘collective nuclear behavior’ are responsible for subsequent LR-asymmetric development of the AMG. In contrast, experiments in genetically modified Drosophila embryos demonstrated that when the nuclei aligned with LR-asymmetry, the subsequent LR-asymmetric development of the AMG was lost. It is known that nuclei are the toughest intracellular organelles. Thus, based on these results, the authors speculated that nuclei may act like structural pillars if they are collectively aligned, which support the structure of the gut and influence LR-asymmetrical changes in shape during its development.
Senior author Kenji Matsuno explains the potential of their findings: “This new method of tracking moving nuclei in vital tissue has helped clarify the role of nuclear alignment in asymmetrical shaping of internal organs during normal development. In addition, our findings are expected to be applied to controlling the shape of regenerative organs. This knowledge could potentially shape future research into organ regeneration which may have applications such as growing artificial organs to model disease mechanisms.”
The article, “Collective nuclear behavior shapes bilateral nuclear symmetry for subsequent left-right asymmetric morphogenesis in Drosophila” was published in Development at DOI: https://doi.org/10.1242/dev.198507.
Related links









